Stanford Researchers Build Sleep ClinicFM: Multimodal Sleep Foundation’s AI Model for 130+ Disease Predictions
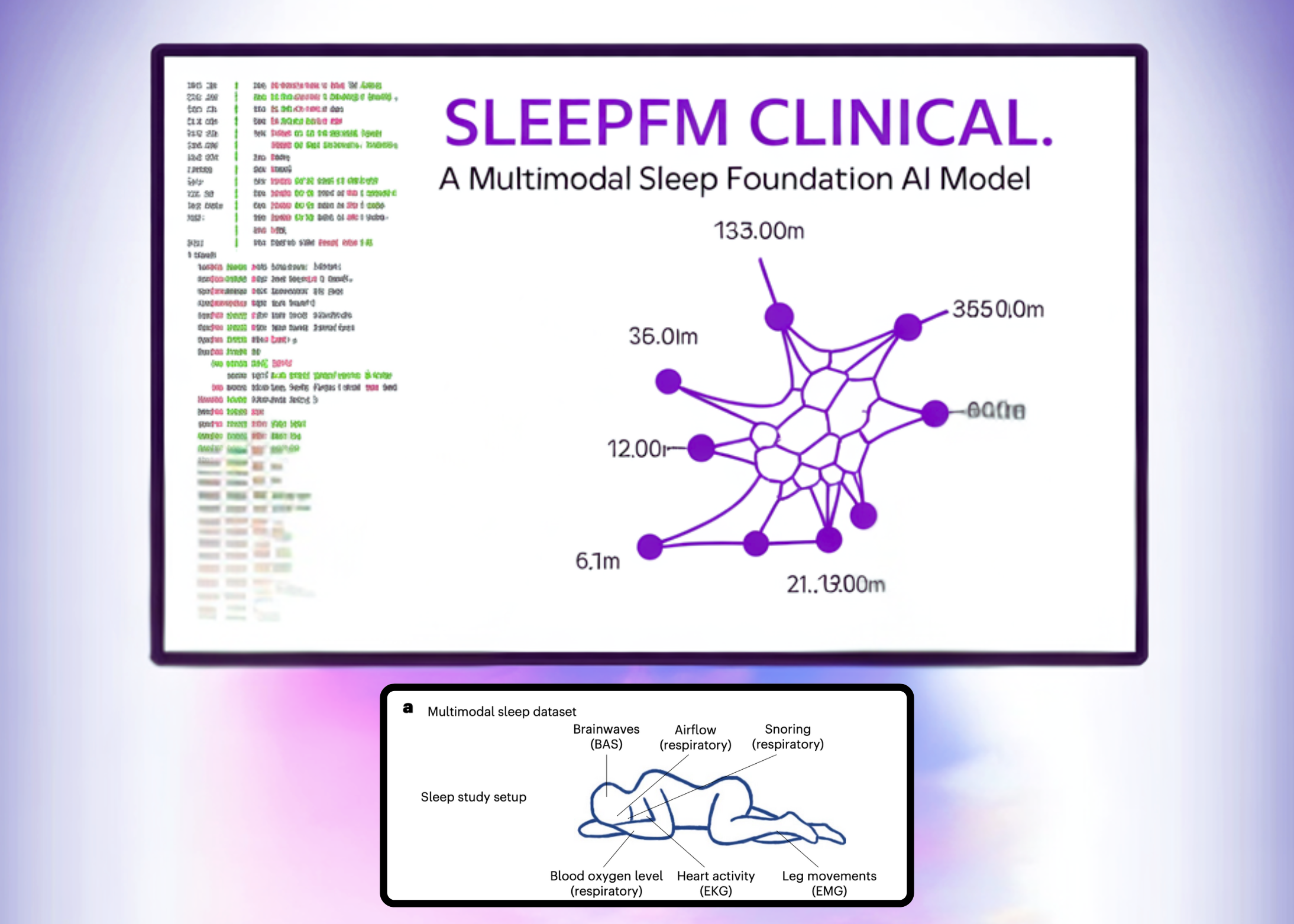
A team of Stanford Medicine researchers has launched SleepFM Clinical, a sleep-based multivariate model that learns from clinical polysomnography and predicts long-term disease risk from a single night of sleep. The research work has been published in Nature Medicine and the team has released the clinical code as open source sleepfm-clinical repository on GitHub under the MIT license.
From nocturnal polysomnography to general representation
Polysomnography records brain activity, eye movements, heart signals, muscle tone, respiratory effort and oxygen saturation during a full night in a sleep lab. It’s a standard gold standard test in sleep medicine, but most clinical workflows only use it for sleep stage and apnea diagnosis. The research team treats these multi-channel signals as a dense time series and trains an underlying model to learn the shared representation across all modes.
SleepFM is trained on approximately 585,000 hours of sleep recordings from approximately 65,000 individuals, taken from multiple cohorts. The largest group comes from the Stanford Sleep Medicine Center, where about 35,000 adults and children had overnight studies between 1999 and 2024. That clinical team is linked to electronic health records, which later enables survival analysis of hundreds of disease categories.
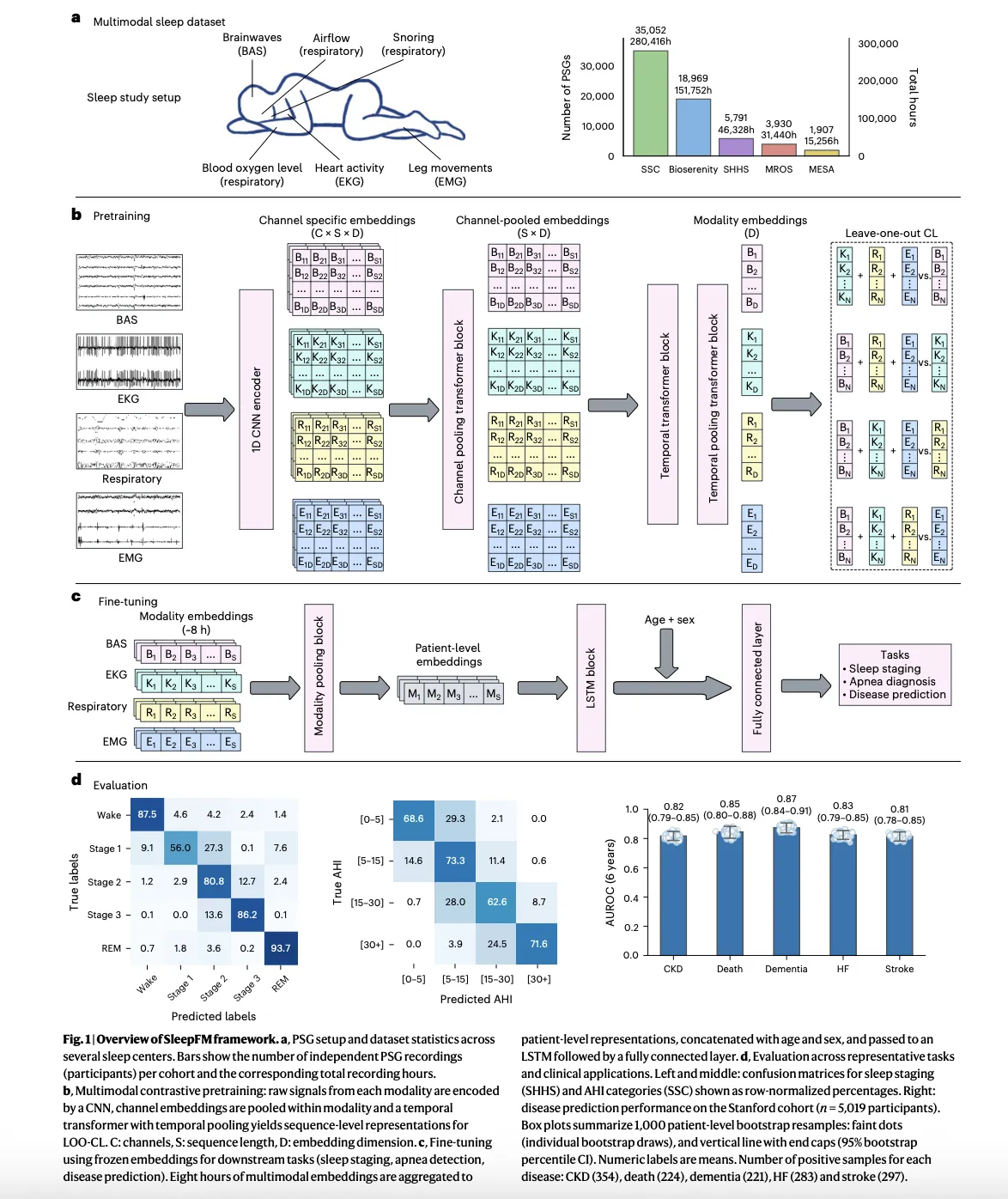
Structural model and purpose of pre-training
At the modeling level, SleepFM uses a convolution core to extract local features from each channel, followed by attention-based clustering across channels and a temporal transform that operates on short segments of the night. The same core structure has already appeared in previous work in SleepFM to find the stage of sleep and the detection of disturbed breathing, where it has been shown that studying the embedding of members in all brain activity, electrocardiography and respiratory signals improves the performance of the river.
The purpose of preparation is to leave out different studies. For each short-term segment, the model builds a separate embedding for each group of mechanisms, such as brain signals, heart signals and respiratory signals, and then learns to synchronize these process embeddings so that any subset predicts the joint representation of the remaining mechanisms. This method makes the model robust to missing channels and different recording montages, common in real-world sleep labs.
After pre-training for off-label polysomnography, the spine is frozen and some special heads are trained. With normal sleep activities, repetitive or vertical lightheadedness indicates embedded sleep stages or apnea labels. To obtain clinical risk prediction, the model aggregates overnights into a single nested patient level, includes basic demographics such as age and sex, and then transfers this expression to a Cox proportional hazards layer for time to event modeling.
Sleep stage benchmarks and apnea
Before moving to disease prediction, the research team confirmed that SleepFM competes with specialized models in common sleep analysis tasks. Previous work has already shown that a simple stage classifier over the SleepFM embedding is very effective in eliminating convolutional networks for sleep stage classification and sleep disordered breathing detection, achieved in macro AUROC and AUPRC in several public datasets.
In a clinical trial, the same pre-trained spine is also used for sleep stage and apnea severity classification across multi-center cohorts. The results reported in the research paper show that SleepFM matches or outperforms existing tools such as traditional evolutionary models and other automated sleep staging systems, ensuring that the presentation captures basic sleep science and not just statistical artifacts from a single dataset.
Predicting 130 illnesses and deaths in one night of sleep
The main contribution of this Stanford research paper is disease prediction. A team of researchers is coding diagnoses in electronic health records at Stanford to enter phecodes and describe more than 1,000 disease groups. For each phecode, they calculated the time to first diagnosis after a sleep study and fit a Cox model over the SleepFM embedding.
SleepFM identifies the effects of 130 diseases whose risk is predicted from a single night of polysomnogram with strong discrimination. This includes all causes of death, dementia, myocardial infarction, heart failure, chronic kidney disease, stroke, atrial fibrillation, many cancers and recurrent cognitive impairment and metabolic disorders. In many of these cases, performance metrics such as concordance index and area under the receiver operating curve are in the range compared to established risk scores, even though the model only uses sleep recordings and basic demographics.
The report also notes that for some cancers, pregnancy complications, circulatory conditions and mental disorders, SleepFM-based predictions reach accuracy rates of nearly 80 percent in multi-year risk windows. This suggests that subtle patterns in the interaction between brain, heart and respiratory signals carry information about subtle disease processes that are not yet clinically apparent.
Comparison with simple bases
To assess the added value, the research team compared SleepFM-based risk models with two baselines. The first uses only demographic characteristics such as age, gender and body mass index. The second one trains the end-to-end model directly on polysomnography and results, without any unsupervised pre-training. Across multiple disease categories, the SleepFM pre-trained representation combined with the light weight head yields higher concordance and a higher long-horizon AUROC than both bases.
This study clearly shows that the benefit comes less from a complex prediction head and more from a basic model that has learned a general representation of sleep physiology. Essentially, this means that clinical centers can reuse a single pre-trained core, read some small site heads with modestly labeled clusters and still approach state of the art performance.
Check it out Paper again FULL CODES here. Also, feel free to follow us Twitter and don’t forget to join our 100k+ ML SubReddit and Subscribe to Our newspaper. Wait! are you on telegram? now you can join us on telegram too.
Check out our latest issue of ai2025.deva 2025-focused analytics platform that transforms model implementations, benchmarks, and ecosystem activity into structured datasets that you can sort, compare, and export
Asif Razzaq is the CEO of Marktechpost Media Inc. As a visionary entrepreneur and engineer, Asif is committed to harnessing the power of Artificial Intelligence for the benefit of society. His latest endeavor is the launch of Artificial Intelligence Media Platform, Marktechpost, which stands out for its extensive coverage of machine learning and deep learning stories that sound technically sound and easily understood by a wide audience. The platform boasts of more than 2 million monthly views, which shows its popularity among viewers.