AI algorithm enables tracking of important white objects | MIT News
The signals that drive the brain and many of the most important functions of the body – consciousness, sleep, breathing, heartbeat, and movement – travel through bundles of “white” fibers in the brainstem, but the thinking systems so far have not been able to properly resolve these important neural networks. That has left researchers and clinicians with little ability to assess how they are affected by trauma or depression.
In a new study, a team of MIT, Harvard University, and Massachusetts General Hospital researchers unveiled AI-powered software that can automatically classify eight different bundles in any MRI sequence.
In an open access study, published Feb. 6 in the Proceedings of the National Academy of Sciences, a research team led by MIT graduate student Mark Olchanyi reports that their BrainStem Bundle Tool (BSBT), which they have made publicly available, has revealed distinct patterns of structural changes in patients with Parkinson’s disease, multiple sclerosis, and traumatic brain injury, and sheds light on Alzheimer’s disease. In addition, the study shows, BSBT enabled repeated tracking of mass healing in a coma patient showing the patient’s seven-month path to recovery.
“The brainstem is an understudied region of the brain because it’s not easy to see,” said Olchanyi, a doctoral candidate in MIT’s Medical Engineering and Medical Physics Program. “People don’t really understand its structure from an image point of view. We need to understand what the organization of white matter is for people and how this organization breaks down in certain problems.”
Adds Professor Emery N. Brown, Olchanyi’s thesis director and senior author of the study, “the brainstem is one of the most important control centers in the body. Mark’s algorithms make an important contribution to cognitive research and to our ability to understand the control of basic physiology. By improving our ability to capture the image of the new respiratory system, he gives the power to control our brain as an important control of the brain, and gives the power to work as a part of the brain that controls the way of breathing systems. of the heart, temperature regulation, how we stay awake during the day and how we sleep at night.”
Brown is the Edward Hood Taplin Professor of Computational Neuroscience and Medical Engineering at the Picower Institute for Learning and Memory, the Institute for Medical Engineering and Science, and the Department of Brain and Psychiatry at MIT. He is also an anesthesiologist at MGH and a professor at Harvard Medical School.
Creating an algorithm
Diffusion MRI helps trace the long branches, or “axons,” of neurons that extend to communicate with each other. Axons are usually coated in a fatty sheath called myelin, and water is dispersed around the axons within the myelin, which is also called the “white matter” of the brain. Diffusion MRI can highlight this highly targeted fluid removal. But isolating the different bundles of axons in the brainstem has been challenging, because they are small and covered by the flow of cerebrospinal fluid and the movements produced by breathing and heartbeat.
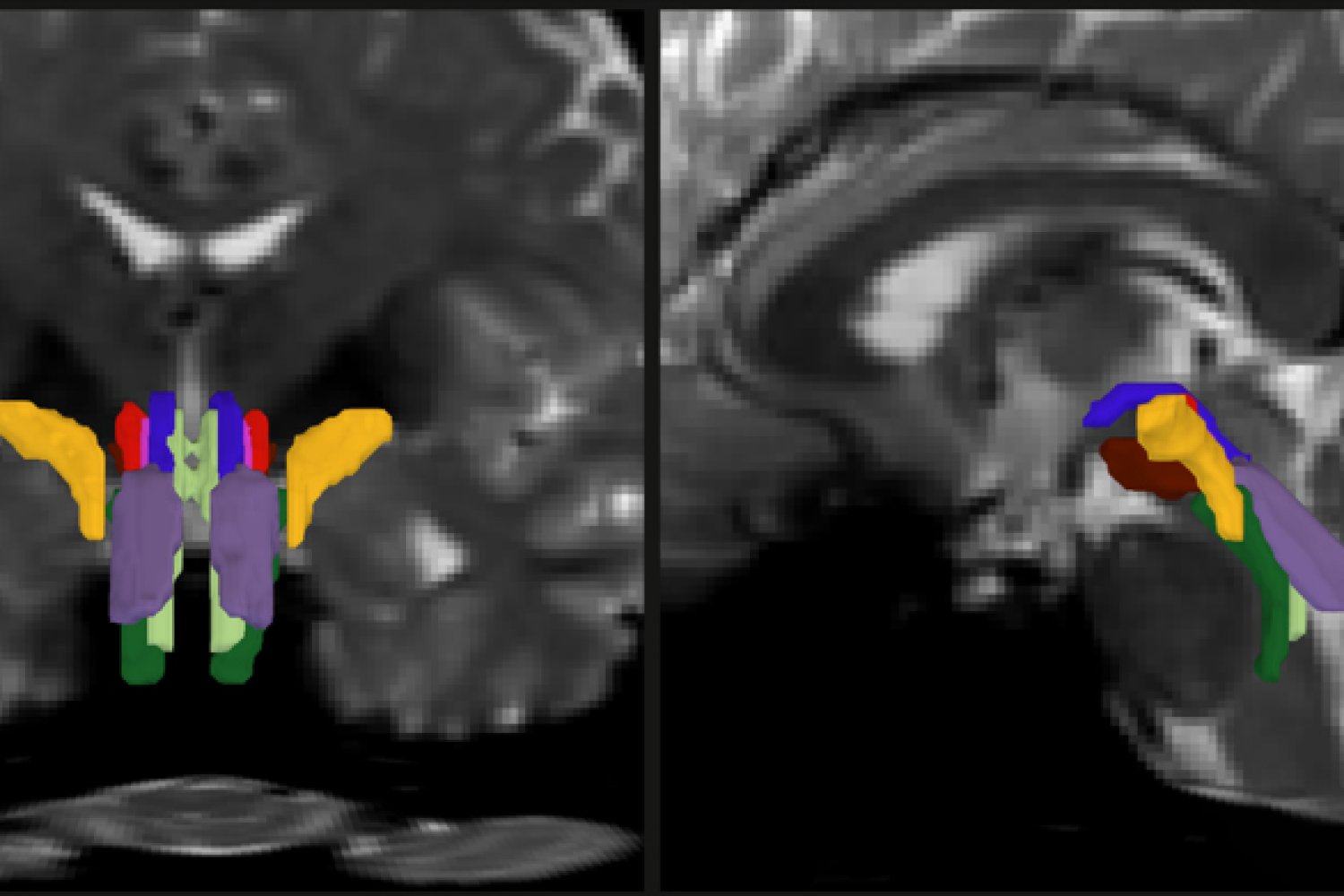
As part of his thesis work to better understand the neural mechanisms that support consciousness, Olchanyi wanted to develop an AI algorithm to overcome these obstacles. BSBT works by tracing fiber bundles entering the brainstem from neighboring higher brain regions, such as the thalamus and cerebellum, to produce a “potential fiber map.” An artificial intelligence module called a “convolutional neural network” then maps and combines several channels of cognitive information from within the brainstem to separate eight individual bundles.
To train the neural network to separate the bundles, Olchanyi “showed” it 30 live-streamed MRI scans from volunteers in the Human Connectome Project (HCP). The scans were manually annotated to teach the neural network how to identify clusters. He then validated the BSBT by testing the output against the “ground truth” of postmortem human brain dissection in which bundles were well defined by microscopic examination or slow but high-resolution imaging. After training, BSBT became proficient at automatically identifying eight different fiber bundles in new scans.
In an experiment to test its consistency and reliability, Olchanyi tasked BSBT with finding bundles from 40 volunteers who were tested separately for two months. In each case, the tool was able to detect the same bundles in the same patients in each of their two scans. Olchanyi also tested BSBT with multiple datasets (not just HCP), and even tested how each part of the neural network contributed to the BSBT analysis by holding them individually.
“We install a neural network using a wringer,” Olchanyi said. “We wanted to make sure that it actually makes these sections that make sense and uses each section in a way that improves accuracy.”
Novel biomarkers are novel
Once the algorithm was properly trained and validated, the research team moved on to test whether the ability to distinguish different fiber bundles in a diffusion MRI scan would allow tracking how the volume and structure of the bundle varies with disease or injury, creating a new type of biomarker. Although the brainstem has been difficult to examine in detail, many studies show that neurodegenerative diseases affect the brainstem, often early in their progression.
Olchanyi, Brown and co-authors applied BSBT to a distributed MRI dataset from patients with Alzheimer’s, Parkinson’s, MS, and traumatic brain injury (TBI). Patients were compared with controls and were sometimes compared with them over time. In the scan, the tool measures bundle volume and “fractional anisotropy,” (FA) which tracks how much water flows in myelinated axons versus how much it spreads in other directions, a proxy for white matter structural integrity.
In each case, the tool found consistent patterns of changes in the bundles. Although only one bundle showed a significant decrease in Alzheimer’s, in Parkinson’s the tool revealed a decrease in FA in three of the eight bundles. It also revealed volume loss in some of the patients between the baseline scan and the two-year follow-up. Patients with MS showed significant FA reduction in four bundles and volume loss in three. Meanwhile, the TBI patients did not show a significant volume decrease in any of the bundles, but a decrease in FA was evident in most of the bundles.
Research studies have shown that the BSBT appears to be more accurate than other methods in differentiating between patients with health conditions compared to controls.
Therefore, BSBT can be “a key adjunct to current diagnostic methods by providing a good structural assessment of brainstem white matter and, in some cases, longitudinal information,” the authors wrote.
Finally, in the case of a 29-year-old man who suffered a severe TBI, Olchanyi applied BSBT to scans taken during seven months of the man’s coma. This device showed that the brain ties of the man were removed, but not cut, and showed that because of his coma, the lesions in the nerve connections decreased by a factor of three in volume. As they healed, the bundles returned to their place.
The authors wrote that BSBT “has significant predictive power for identifying preserved brain bundles that may facilitate recovery from coma.”
The other senior authors of the study are Juan Eugenio Iglesias and Brian Edlow. Other co-authors are David Schreier, Jian Li, Chiara Maffei, Annabel Sorby-Adams, Hannah Kinney, Brian Healy, Holly Freeman, Jared Shless, Christophe Destrieux, and Hendry Tregidgo.
Funding for this research comes from the National Institutes of Health, the US Department of Defense, the James S. McDonnell Foundation, the Rappaport Foundation, the American SidS Institute, the American Brain Foundation, the American Academy of Neurology, the Center for Integration of Medicine and Innovative Technology, the Blueprint for Neuroscience Research, and the Massachusetts Life Sciences Center.